library(tidyverse)
googlecrs <- "EPSG:4326"
localUTM <- "EPSG:32615"
path <- "/home/ajackson/Dropbox/Rprojects/TexasDioceseCreationCare/Data/"Read and Analyze subsidence data
Setup
Data scraped (with some difficulty) from https://www.arcgis.com/home/webmap/viewer.html?webmap=1e3c97ed53e2476bb842ceccd6a90514&extent=-96.3605,29.2149,-94.1042,30.3246
Found from the website for the Harris-Galveston Subsidence District.
Scraping process
Install clipit to increase clipboard buffer size
Open table
Open information screen
Find table in info screen
Select all the data with
ctl-CDump into file with
xsel -b > foo8Shorten extremely long line with
cat foo8 | sed 's/<tr>/\r<tr>/g' > foo9.htmlRead into editor and add
<html> <body> <table>after eliminating un-needed stuff.
Read and parse the data
# Import the data
df <- rvest::read_html(paste0(path, "SubsidenceDataHarris.html")) %>%
rvest::html_table()
df <- do.call(rbind, df)
df <- df %>%
rename(Station=X1, Operator=X2, Latitude=X3, Longitude=X4, Start_Year=X5, End_Year=X6,
Years_Monitoring=X7, Total_Vertical_Displacement_cm=X8, Subsidence_Rate_cmperyr=X9,
POR_Plot=X10, RateLabel=X11, FID=X12
)
# Some records are recorded at stations at the same location, which screws up kriging.
# We will find those records and replace them with one record at that spot and average
# the variable values.
df <- df %>%
mutate(index=paste0(as.character(Latitude), as.character(Longitude))) %>%
group_by(index) %>%
summarise(
Station=first(Station),
Operator=first(Operator),
Subsidence_Rate_cmperyr=mean(Subsidence_Rate_cmperyr),
Total_Vertical_Displacement_cm=mean(Total_Vertical_Displacement_cm),
Start_Year=min(Start_Year),
End_Year=max(End_Year),
Years_Monitoring=max(Years_Monitoring),
NumInAvg=n(),
Latitude=first(Latitude),
Longitude=first(Longitude)) %>%
select(-index)
df_sf <- sf::st_as_sf(df, coords=c("Longitude", "Latitude"), crs=googlecrs, agr = "identity")
df_xy <- sf::st_transform(df_sf, crs=localUTM)
# saveRDS(df_xy, paste0(path, "Subsidence_xy.rds"))
bbox <- sf::st_bbox(df_sf)
# Expand box by 20% to give a little extra room
Dx <- (bbox[["xmax"]]-bbox[["xmin"]])*0.1
Dy <- (bbox[["ymax"]]-bbox[["ymin"]])*0.1
bbox["xmin"] <- bbox["xmin"] - Dx
bbox["xmax"] <- bbox["xmax"] + Dx
bbox["ymin"] <- bbox["ymin"] - Dy
bbox["ymax"] <- bbox["ymax"] + Dy
bb <- c(bbox["xmin"], bbox["ymin"], bbox["xmax"], bbox["ymax"])Plot the data
Base_basemapR <- basemapR::base_map(bbox, basemap="mapnik", increase_zoom=2)
# This is the best looking one.
df_sf %>%
ggplot() +
Base_basemapR +
geom_sf()
Grid and contour
Though really what I care most about is the grid, since that will yield the subsidence values at each of the church locations
# First we create a box to build the grid inside of - in XY coordinates, not
# lat long.
bbox_xy = bbox %>%
sf::st_as_sfc() %>%
sf::st_transform(crs = localUTM) %>%
sf::st_bbox()
# sfc POINT file
grd_sf <- sf::st_make_grid(x=bbox_xy,
what="corners",
cellsize=2000,
crs=localUTM
)
# Data frame
grd_df <- expand.grid(
X = seq(from = bbox_xy["xmin"], to = bbox_xy["xmax"]+2000, by = 2000),
Y = seq(from = bbox_xy["ymin"], to = bbox_xy["ymax"]+2000, by = 2000) # 1000 m resolution
)Inverse distance weighting
# Create interpolator and interpolate to grid
fit_IDW <- gstat::gstat(
formula = Subsidence_Rate_cmperyr ~ 1,
data = df_xy,
#nmax = 10, nmin = 3, # can also limit the reach with these numbers
set = list(idp = 2.5) # inverse distance power
)
# Use predict to apply the model fit to the grid (using the data frame grid
# version)
# We set debug.level to turn off annoying output
interp_IDW <- predict(fit_IDW, grd_sf, debug.level=0)
# Convert to a stars object so we can use the contouring in stars
interp_IDW_stars <- stars::st_rasterize(interp_IDW %>% dplyr::select(Subsidence_Rate_cmperyr=var1.pred, geometry))
# Quick sanity check, and can use to adjust the distance power Looks pretty
# good - most input points look close to the output grid points, with some
# notable exceptions. The red point to the north is possibly bad data. Easier
# to judge from areal displays.
ggplot() +
# geom_stars is a good way to display a grid
stars::geom_stars(data=interp_IDW_stars) +
geom_sf(data=df_xy, aes(color=Subsidence_Rate_cmperyr), size=5) +
geom_sf(data=df_xy, color="Black", size=1) +
# This is for the raster color fill
scale_fill_gradientn(colors=rainbow(5), limits=c(-2,1)) +
# This is for the original data points
scale_color_gradientn(colors=rainbow(5), limits=c(-2,1)) +
labs(title="Inverse Distance Weighting, Power = 2.5")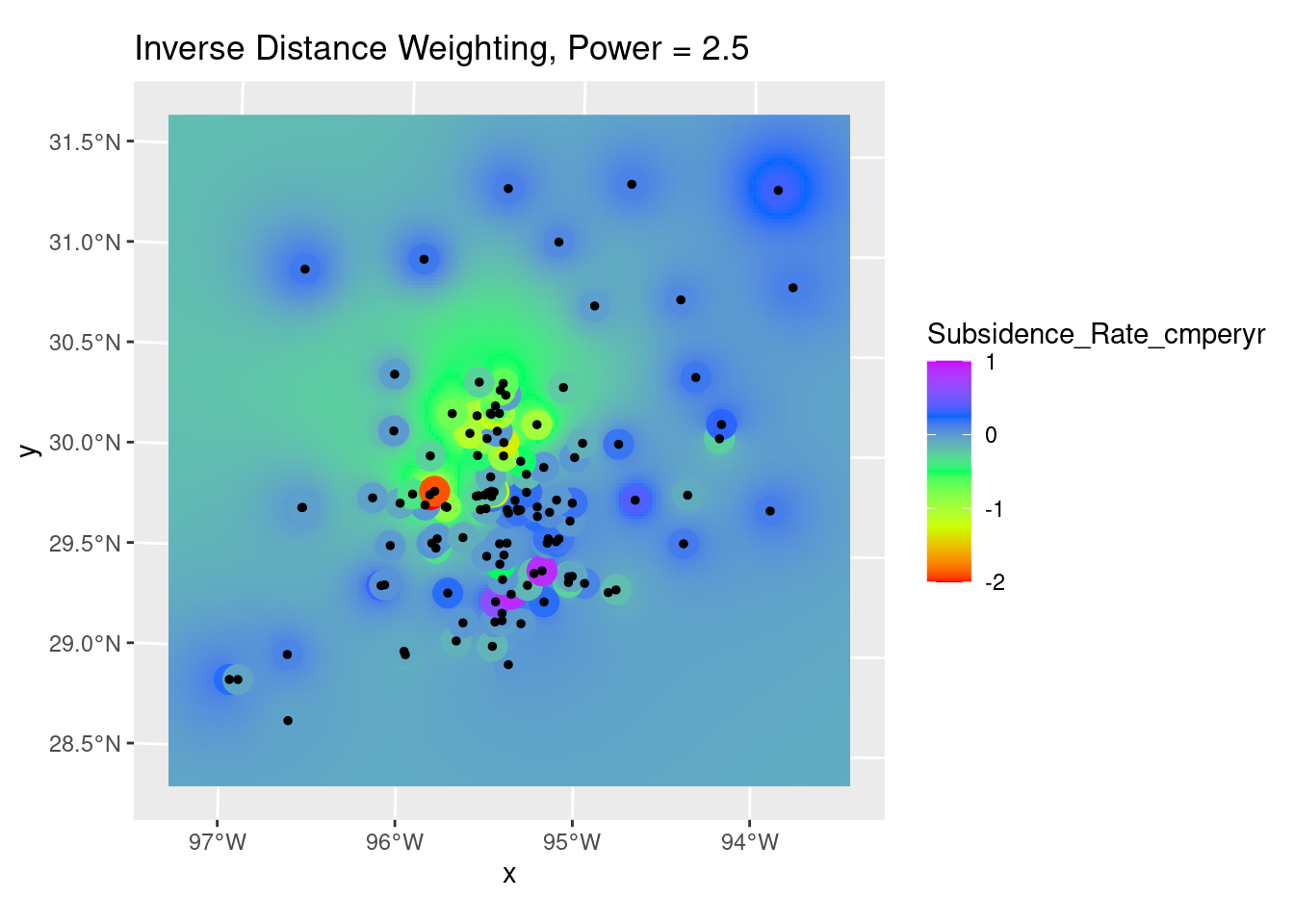
Search for best IDW power
Even optimizing, the result is kind of sucky.
# Do a leave-one-out analysis for a variety of weighting powers
Validate <- NULL
for (power in (2:8)*0.5) {
my_fit <- gstat::gstat(formula = Subsidence_Rate_cmperyr ~ 1, data = df_xy, set = list(idp = power))
foo <- sf::st_as_sf(gstat::gstat.cv(my_fit, debug.level=0, verbose=FALSE)) %>%
rename(Observed=observed, Predicted=var1.pred ) %>%
mutate(power=power)
Validate <- bind_rows(Validate, foo)
}
Validate %>%
ggplot(aes(x=Observed, y=Predicted)) +
geom_point() +
geom_smooth(method='lm') +
geom_abline(intercept=0, slope=1, color="red", linetype="dashed") +
facet_wrap(vars(power)) +
labs(title="Leave-one-out Validation",
x="Observed (inches)",
y="Predicted (inches)")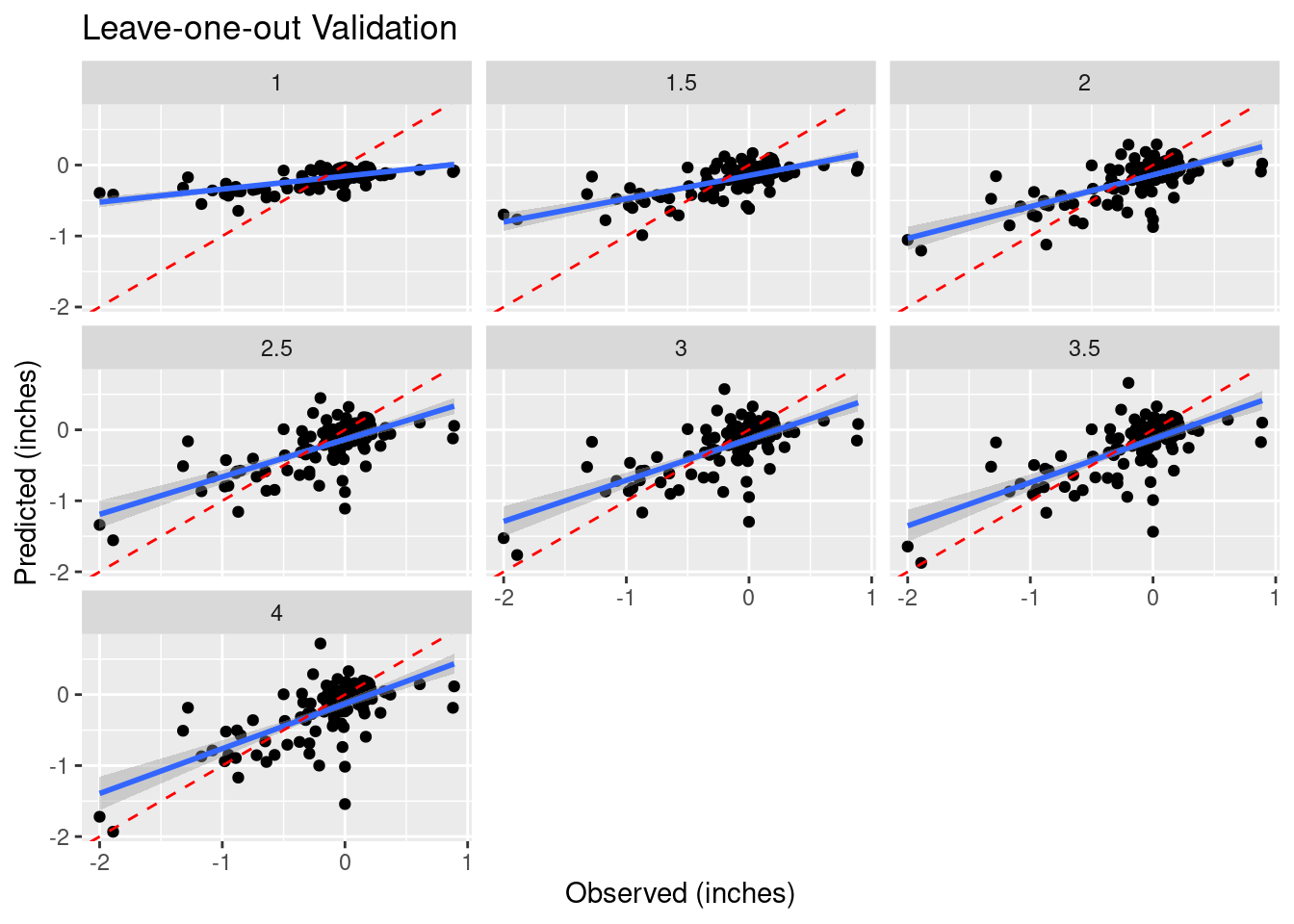
# Root mean square error
RMS <- Validate %>%
group_by(power) %>%
summarise(RMS=sqrt(sum((Predicted-Observed)^2) / n()))
RMS %>%
ggplot(aes(x=power, y=RMS)) +
geom_point() +
geom_smooth() +
labs(title="RMS Error vs. Inverse Distance Weighting Power")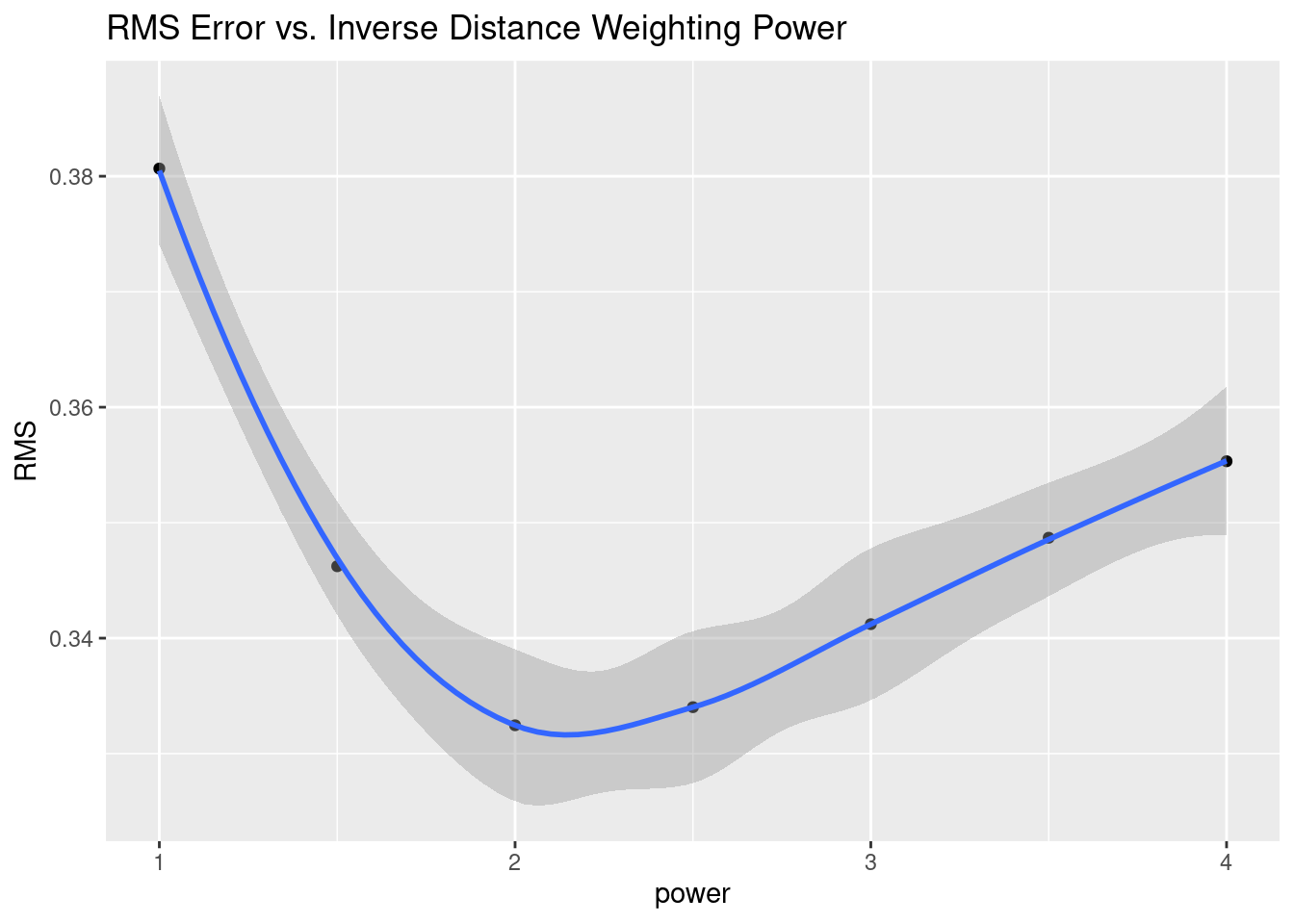
Kriging
First the drift.
I don’t like the looks of the fit - I think that there is enough data surrounding the bowl near zero that there is no drift. This is a flat plane with a bowl in the middle.
# Let's look at 1st order fit
df_xy_df <- bind_cols(sf::st_drop_geometry(df_xy),
sf::st_coordinates(df_xy)) # make a tibble
# Define the 1st order polynomial equation
f.1 <- as.formula(Subsidence_Rate_cmperyr ~ X + Y)
# Run the regression model
lm.1 <- lm( f.1, data=df_xy_df)
# Use predict to apply the model fit to the grid This re-attaches X-Y
# coordinates to the predicted values so they can be made into a grid
Poly_fit.1 <- sf::st_as_sf(bind_cols(grd_df,
data.frame(var1.pred=predict(lm.1, grd_df))),
crs=localUTM,
coords=c("X", "Y"))
# Convert to a stars object
Poly_fit_star.1 <- stars::st_rasterize(Poly_fit.1 %>%
dplyr::select(Subsidence_Rate_cmperyr=var1.pred, geometry))
ggplot() +
stars::geom_stars(data=Poly_fit_star.1) +
scale_fill_gradientn(colors=rainbow(5)) +
geom_sf(data=df_xy, aes(color=Subsidence_Rate_cmperyr), size=5) +
scale_color_gradientn(colors=rainbow(5), limits=c(-2,1)) +
scale_fill_gradientn(colors=rainbow(5), limits=c(-2,1)) +
geom_sf(data=df_xy, color="Black", size=1)+
labs(title="Linear trend from subsidence data")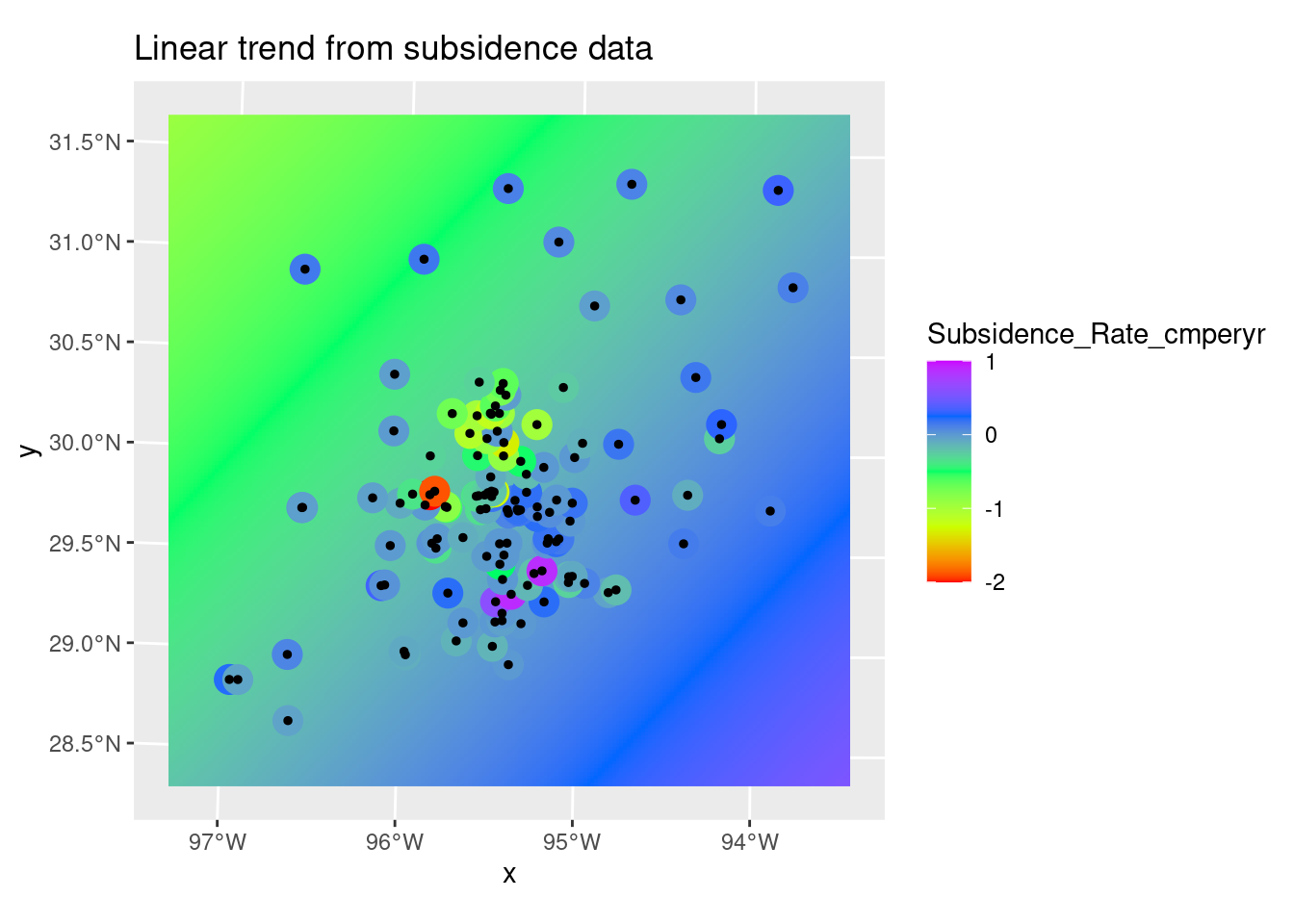
Kriging
I pick the Spherical model
# Let's look at the variogram
var.smpl <- gstat::variogram(Subsidence_Rate_cmperyr ~ 1,
data=df_xy)
# Compute the variogram model by passing the nugget, sill and range values
# to fit.variogram() via the vgm() function.
# models are "Sph", "Exp", "Mat", and "Gau"
Plot_data <- NULL
for (model in c("Sph", "Exp", "Mat", "Gau")) {
foo <- gstat::fit.variogram(var.smpl, fit.ranges = TRUE, fit.sills = TRUE,
debug.level = 0,
gstat::vgm(psill=NA, model=model, range=NA, nugget=0))
Plot_data <- bind_rows(Plot_data, gstat::variogramLine(foo, maxdist = max(var.smpl$dist)) %>%
mutate(Model=model))
}
ggplot(var.smpl, aes(x = dist, y = gamma)) +
geom_point() +
geom_text(aes(hjust="left", label=np), nudge_x=200) +
geom_line(data=Plot_data, aes(x=dist, y=gamma, color=Model)) +
labs(title="Variogram for detrended data",
subtitle="Numbers represent number of points in the distance bin")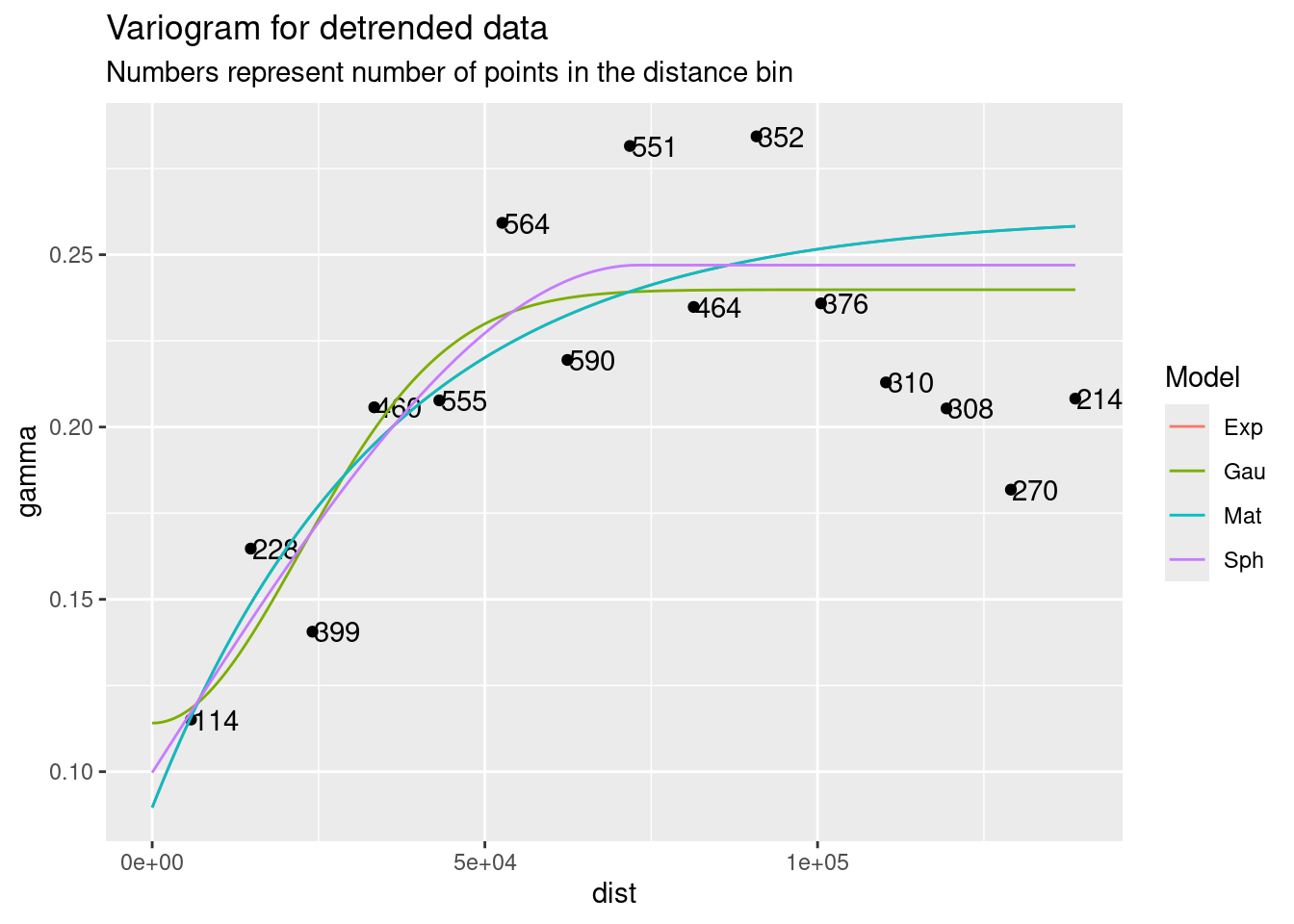
Do Kriging
# We have our model, so now let's use it
dat.fit <- gstat::fit.variogram(var.smpl, fit.ranges = TRUE, fit.sills = TRUE,
debug.level = 0,
gstat::vgm(psill=NA, model="Gau", range=NA, nugget=0))
# saveRDS(dat.fit, paste0(path, "gstat_variogram_Subsidence.rds"))
Krigged_grid <- gstat::krige(Subsidence_Rate_cmperyr ~ 1,
df_xy,
grd_sf,
debug.level=1,
dat.fit)[using ordinary kriging]Krigged_star <- stars::st_rasterize(Krigged_grid %>% dplyr::select(Subsidence_Rate_cmperyr=var1.pred, geometry))
# Residuals
ggplot() +
stars::geom_stars(data=Krigged_star) +
scale_fill_gradientn(colors=rainbow(5)) +
geom_sf(data=df_xy, color="Black", size=1) +
labs(title="Residuals from Kriging")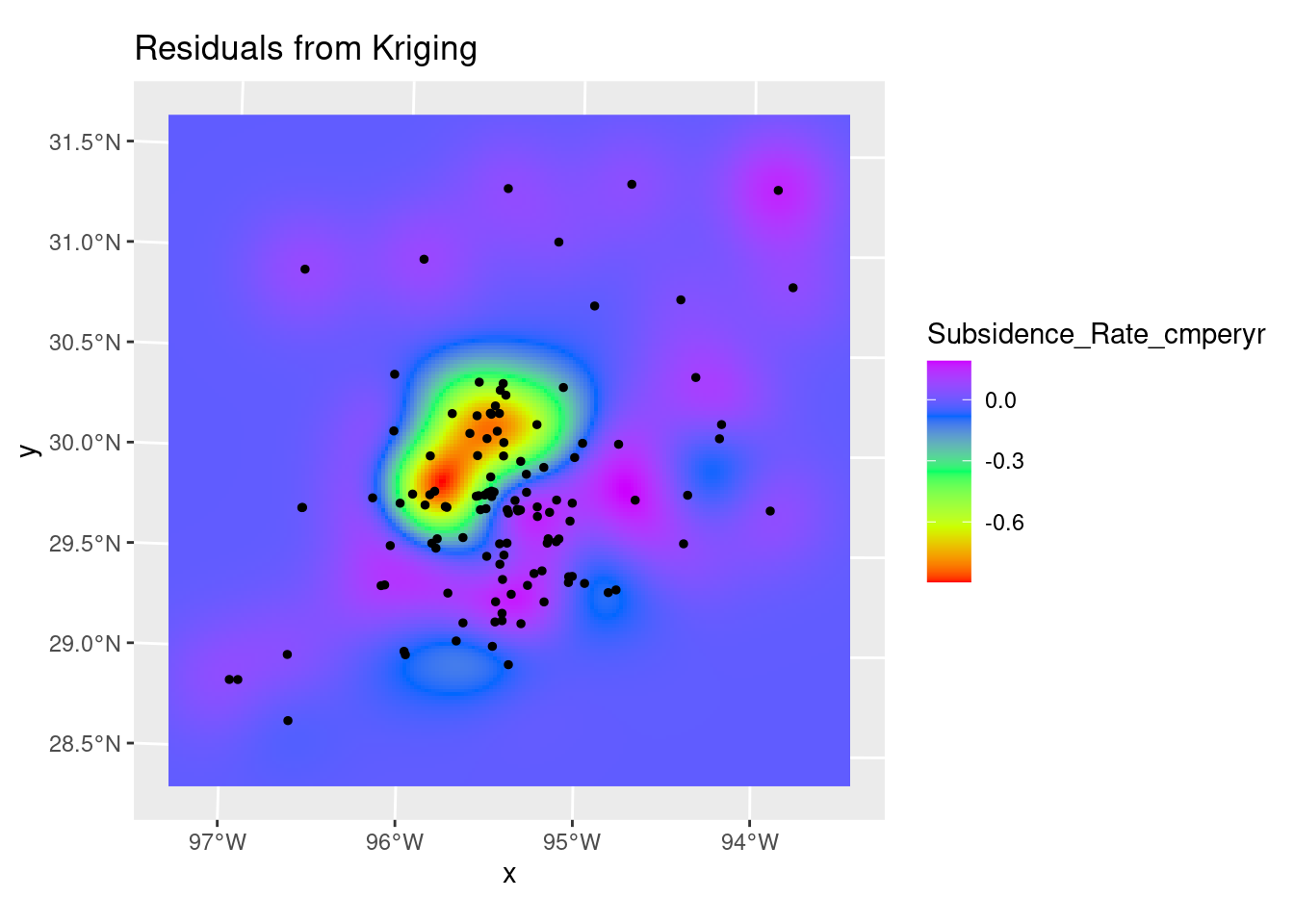
Contours
brks <- seq(from = -1, to = 0.2, by = 0.2)
# Create contour lines
Contours_Krig <- stars::st_contour(Krigged_star, contour_lines=TRUE, breaks=brks)
# Plot to see what it all looks like
ggplot() +
stars::geom_stars(data=Krigged_star) +
scale_fill_gradientn(colors=rainbow(5), limits=c(-1,0.2)) +
geom_sf(data=df_xy, aes(color=Subsidence_Rate_cmperyr), size=5) +
scale_color_gradientn(colors=rainbow(5), limits=c(-1,0.2)) +
geom_sf(data=df_xy, color="Black", size=1) +
geom_sf(data=Contours_Krig, color="black") 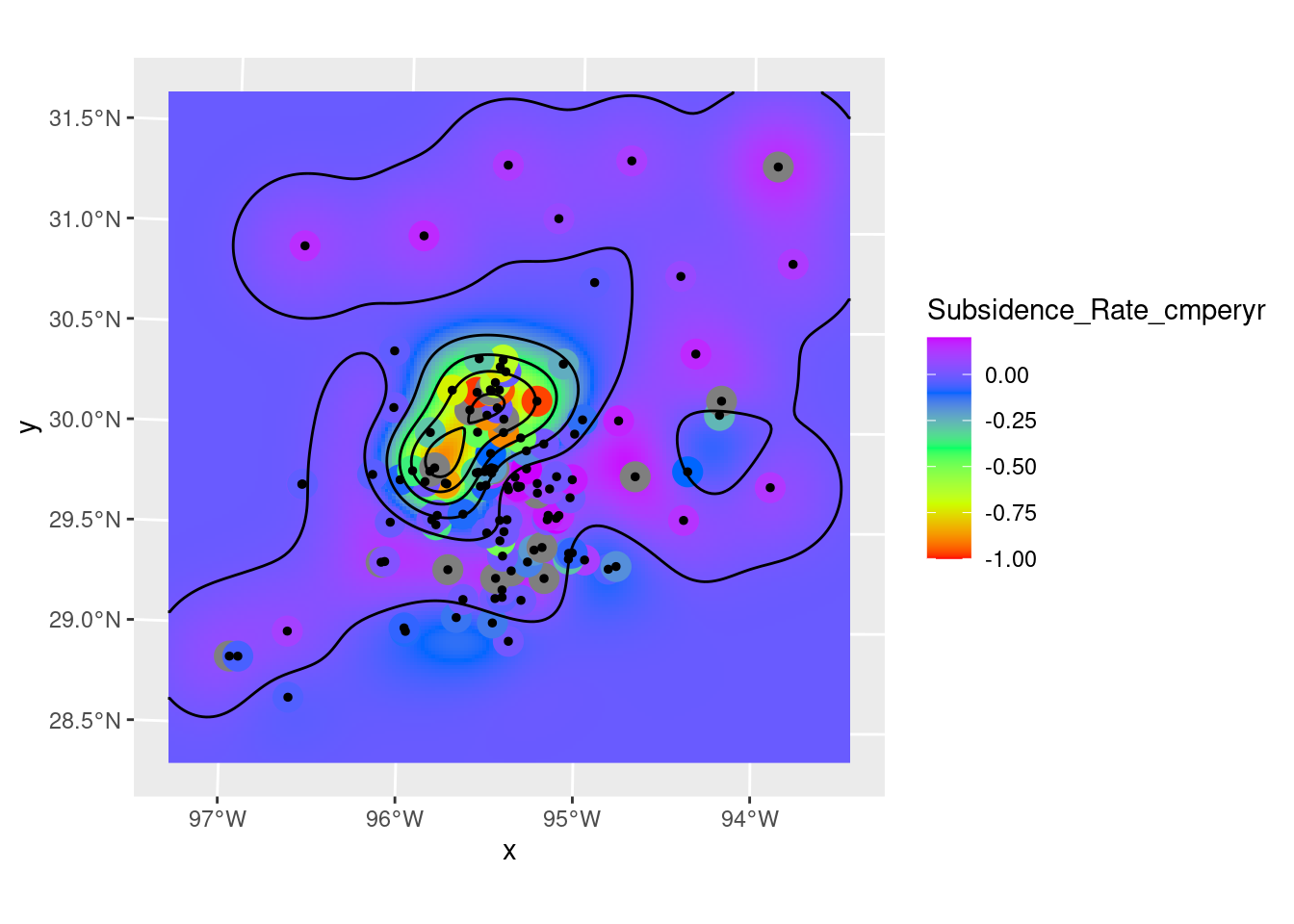
Final publication quality map
# Now lets transform the contours to lat-long space and plot on the basemap
Contours_LL <- sf::st_transform(Contours_Krig, crs=googlecrs)
Krig_star_LL <- sf::st_transform(Krigged_star, crs=googlecrs)
# saveRDS(Contours_LL, paste0(path, "Contours_LL_Subsidence.rds"))
# saveRDS(Krig_star_LL, paste0(path, "Star_Grid_LL_Subsidence.rds"))
ggplot() +
# this is the best looking basemap
Base_basemapR +
# Gridded data
stars::geom_stars(data=Krig_star_LL, alpha=0.4) +
# Add points
geom_sf(data=df_sf) +
# Create filled density "contours"
geom_sf(data=Contours_LL, color="black") +
scale_fill_viridis_c(direction=-1, alpha=0.4, name="Subsidence\nRate (cm/yr)") +
# Add a scale bar
ggspatial::annotation_scale(data=df_sf, aes(unit_category="imperial", style="ticks"),
location="br", width_hint=0.2, bar_cols=1) +
# Add CI annotation at specified window coordinates
annotate("text", label="C.I. = 0.2 cm per year subsidence
Data sourced from the Harris-Galveston Subsidence District
2022 Annual Groundwater Report
Map a product of Alan Jackson",
x=-Inf,
y=-Inf,
hjust="inward", vjust="inward", size = 2) +
coord_sf(crs=googlecrs) + # required
# Add title
labs(title="Subsidence Rate",
subtitle="Points represent the measurement locations",
x="Longitude", y="Latitude")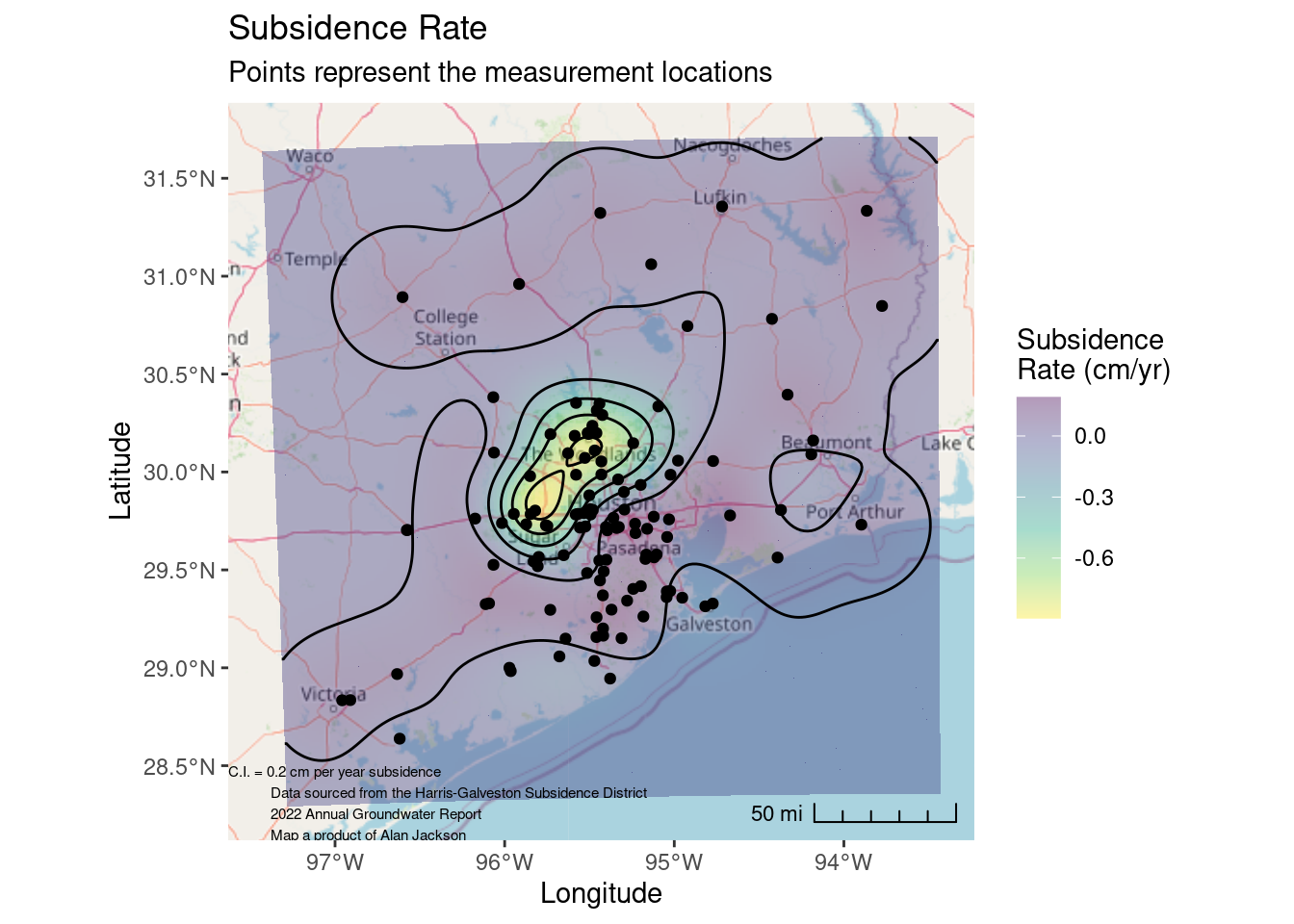
# ggsave(paste0(path, "Subsidence_map.png"),dpi=400)